Howdy!
This week was pretty interesting because I was able to see all of the electrospun fibers we created last week with using the scanning electron microscope (SEM). I was also able to practice my students will use to measure the contact angle of a drop of water on an object to test its level of hydrophobicity.
If you recall from my earlier blogs, we want to create membrane that is superhydrophobic. According to all of the background research, superhydrophobic surfaces have a a high water contact angle above 150° and a low water roll off angle below 10°.
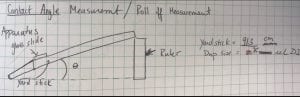
I didn’t take a picture of our set up, so I’ll explain it. It’s something that can be modified easily and done in class. We taped a glass slide to a yard stick and then mounted our membrane sample with double sided tape. We then dropped 75µL of water on the membrane. Next, we very slowly raised our meter stick against the wall . We measured the height we raised the ruler up the wall when the drop of water rolled off the sample. We used trigonometry to figure out the roll off angel, but if you have protractors you could have your students measure the angle directly. Here is our set up to test roll off angle.

After testing roll off angle, we were able to use the drop shape analyzer, which allowed us to take a precise measurement of the contact angle between a drop of water and our membrane. Here you can really see the different wetting properties of the two membranes.
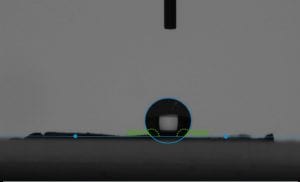

This is also something I can do with my students. I would have them take a picture of their drop of water and then after uploading it into an image editor they can measure and label their contact angles.
Next, we took our samples to the SEM to look at the surface morphology.



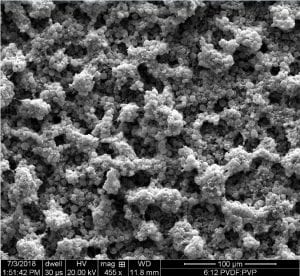
Overall our our membrane with only PVDF and our membrane with 6:12 PVDF: PVP had the highest contact angle. The PVDF only membrane had no roll off angle and the drop of water adhered to the membrane, but the 6:12 had our lowest roll off data.
Based on our data, we decided to blend our best membranes. We will spin fibers that have 18:0 PVDF:PVP inner core and an 6:12 PVDF:PVP outer shell.

Next steps include adding silver nanoparticles to the membrane to add roughness and then coating the membrane with a chemical to lower the surface energy. So I will update you then.
Until next time…