In my lab we are working with a virus called AAV9. They are currently using as a vector for gene therapy, specifically to the heart. The virus is currently able to identify inflammation but the goal is to make it specific to a cell. To do this, my mentor is working with adding locks that will allow for the virus to only attach to a specific type of cell. This is very challenging and there are many steps to the process- from the creation of a virus with specific locks to the actual delivery of the virus- in vitro or in vivo.
My job in the lab currently involves keeping my HEK 293T cells alive- this involves splitting the cells: lifting them, changing them to another plate and adding new media to keep them happy. This is the one process that I feel very comfortable doing by myself and my mentor has trusted me to do it already about 3 times without his supervision.. yay! The rest of the time my mentor has been teaching me new protocols with the end goal of being able to make virus! Last week we started by preparing the backbone and the inserts that we wanted in our plasmid. Once the plasmid is ready, we utilize it to transform E. coli. We then send our E.coli samples to a company that is in charge of sequencing them and checks to see if the bacteria did indeed take in the plasmid. We sent in 8 samples and 4 out of the 8 were great! This allowed us to go on to what we have been working on this week- Maxi Prepping our DNA.
A Maxi Prep works as follows: Making large amounts of bacteria, spinning down the bacteria to a pellet –> lysing the bacteria to release DNA and protein –> precipitating the protein –> get rid of the protein and load DNA solution onto column that can capture DNA –> elute DNA and precipitate DNA –> pellet DNA and redissolve DNA.
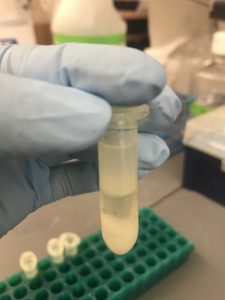
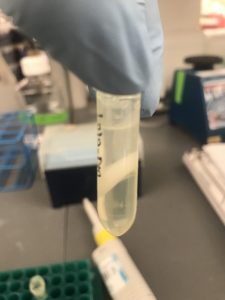
What you see there inside the bottle is the pellet which is part of the first step in a Maxi Prep- I love pipetting but never have I had to spend a good 3 -5 minutes trying to re-suspend pellets– My thumb is definitely getting muscular! After that all the steps to follow involve cleaning the DNA because we want to make sure that we don’t have any cell debris or RNA in our final products. The pictures below show some of the cleaning/precipitation steps used in cleaning DNA. Picture on the left has a layer of gel, DNA and phenol/chloroform/isoamyl alcohol, After spinning them, the layers rearrange as the picture on the right with the layer of the phenol/chloroform/isoamyl alcohol on the bottom, gel and then the DNA on top which is what we will extract.
After all the cleaning and precipitation steps are done we will carry out a gel to see if we have clean DNA we can then use for our virus!

That’s great that your mentor is trusting you so much! I’m having to do a lot of pipetting as well, so I’m glad that someone else is sharing that experience.
I feel very nervous when I’m flying solo but my mentor is really good about giving specific instructions and also has me go over them before he leaves me to work on my own. I really appreciate that because it takes so long to do some stuff and you can’t really check if you did it right until the end. I find myself always asking how things are supposed to look Iike if I do something incorrectly. Once the results come out and everything looks well I feel so relieved.
You look like you are doing some really cool stuff! I’m kind of jealous I’m not working so much in a lab – but I do like the research I am doing. That’s great that you’ve been doing this on your own!
This is so super cool!! As you have already commented, we are both using viruses in our research and working on modifying them for specific purposes, but the techniques we are using are so different! I would love to learn more from you about the genetic engineering technique of modifying viruses. It would be awesome if I could incorporate some of that into my lesson. Great work!